We’re excited to announce the publication of The Plant Cell’s latest Teaching Tool, “Computational Image Processing in Microscopy,” by Adrienne Roeder, available without subscription at Plantae.org (and in the October issue of The Plant Cell).
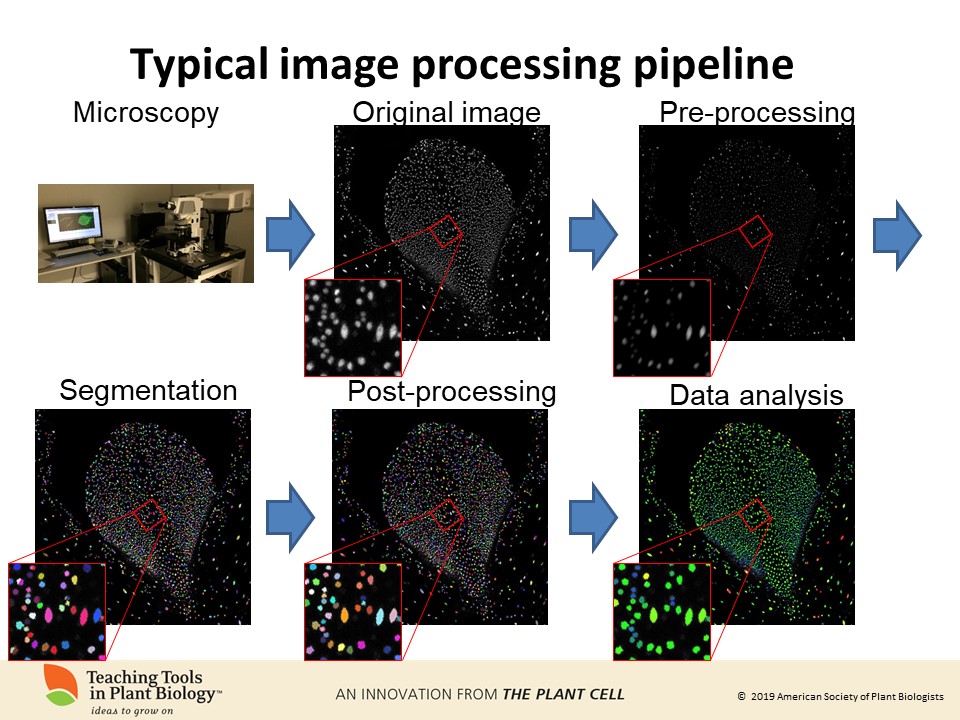
 The age of big data includes sophisticated imaging datasets. Computational image processing is essential for extracting quantitative information from these large image datasets. Computer scientists have been working for decades to build image analysis tools. It is critical for biologists to understand the concepts in image processing so that they can communicate with computer scientists in designing image processing pipelines and applying these tools to their own images.
The age of big data includes sophisticated imaging datasets. Computational image processing is essential for extracting quantitative information from these large image datasets. Computer scientists have been working for decades to build image analysis tools. It is critical for biologists to understand the concepts in image processing so that they can communicate with computer scientists in designing image processing pipelines and applying these tools to their own images.
We focus on microscopy images, but the principles apply to other types of images as well. Furthermore, it is important to understand what manipulations are appropriate in preparing images for publication, what manipulations must be disclosed in the methods and the figure legends, and what manipulations are unacceptable.
Here we introduce computational image analysis concepts and terms and illustrate them with Fiji and the COSTANZA (COnfocal STack ANalyZer) plugin. We provide a step by step, hands-on workshop with a sample image so that students can try some of these functions themselves. Additional image files are also provided for further exploration.
 Dr. Adrienne Roeder, Associate Professor at Cornell University, has had a longstanding interest in combining biology with computation since she was an undergraduate student at Stanford where she majored in Biology with a minor in Mathematical and Computational Science. She received her PhD from UC San Diego in 2005 studying fruit development and seedpod dehiscence in Arabidopsis. Then she was a postdoctoral scholar at Caltech where she was involved in establishing the computational morphodynamics approach to understanding morphogenesis by combining live imaging, image processing, and computational modeling. Now she is the Nancy M. and Samuel C. Fleming Term Associate Professor in the Weill Institute for Cell and Molecular Biology and the School of Integrative Plant Science, Section of Plant Biology at Cornell University. Her lab uses a computational morphodynamics approach to study how cell sizes and organ sizes are controlled in Arabidopsis. Through this research, the themes emerging are the importance of stochasticity and heterogeneity at the cellular level which the plant utilizes to develop robust organs with the correct size, shape, and functions.
Dr. Adrienne Roeder, Associate Professor at Cornell University, has had a longstanding interest in combining biology with computation since she was an undergraduate student at Stanford where she majored in Biology with a minor in Mathematical and Computational Science. She received her PhD from UC San Diego in 2005 studying fruit development and seedpod dehiscence in Arabidopsis. Then she was a postdoctoral scholar at Caltech where she was involved in establishing the computational morphodynamics approach to understanding morphogenesis by combining live imaging, image processing, and computational modeling. Now she is the Nancy M. and Samuel C. Fleming Term Associate Professor in the Weill Institute for Cell and Molecular Biology and the School of Integrative Plant Science, Section of Plant Biology at Cornell University. Her lab uses a computational morphodynamics approach to study how cell sizes and organ sizes are controlled in Arabidopsis. Through this research, the themes emerging are the importance of stochasticity and heterogeneity at the cellular level which the plant utilizes to develop robust organs with the correct size, shape, and functions.
Adrienne writes, “I love collaborating with scientists from other disciplines to answer challenging biological questions. To collaborate effectively, we all have to learn each other’s specialized languages. As a postdoc, I was introduced to computational image processing for the first time, and I could see it was what I needed to answer the question of how giant cells formed among small cells in the sepal, but I struggled with picking up the language of image processing. To help myself, when I encountered a term I did not know, I wrote it down and looked it up. Over time, I wrote an annotated glossary of the image analysis terms. I decided that if I needed the glossary probably it would be helpful to other biologists, so I published it. With this teaching tool, I hope to further lower the barrier between biologists using computational image processing to analyze their images, by pairing the image processing concepts with a hands-on workshop using freely available image processing software and plugins.”

We thank you for all your efforts in editing the articles.
We wish you more progress in your projects that you publish on the web pages.