Review: Wheat genomics comes of age
 Due to its highly repetitive, polyploid genome, wheat genomics has lagged behind that of other cereals, but new tools promise to begin closing that gap. Uauy reviews these new tools, which include access to full genomes of several wheat varieties, gene expression data from hundreds of publicly available RNA-sequencing datasets, next-generation sequencing enabled trait mapping, and genome editing approaches. He concludes with a call to action: “It is essential that we train the next generation of genomics-enabled crop researchers and breeders. This does not necessarily imply that a single individual be able to move seamlessly from the field to the computer; but rather that they be able to communicate effectively between disciplines and appreciate the potential of genomics and field-based research to complement each other.” Curr. Opin. Plant Biol. 10.1016/j.pbi.2017.01.007 Tags: Applied Plant Biology, Computational Biology, Genetics, Genomics
Due to its highly repetitive, polyploid genome, wheat genomics has lagged behind that of other cereals, but new tools promise to begin closing that gap. Uauy reviews these new tools, which include access to full genomes of several wheat varieties, gene expression data from hundreds of publicly available RNA-sequencing datasets, next-generation sequencing enabled trait mapping, and genome editing approaches. He concludes with a call to action: “It is essential that we train the next generation of genomics-enabled crop researchers and breeders. This does not necessarily imply that a single individual be able to move seamlessly from the field to the computer; but rather that they be able to communicate effectively between disciplines and appreciate the potential of genomics and field-based research to complement each other.” Curr. Opin. Plant Biol. 10.1016/j.pbi.2017.01.007 Tags: Applied Plant Biology, Computational Biology, Genetics, Genomics
Review: Mechanisms to mitigate the tradeoff between growth and defense ($)
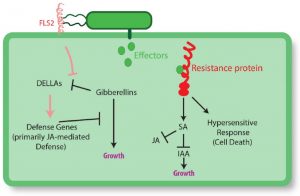 It is widely recognized that defense incurs a cost in terms of reduced growth. Karasov et al. explore the nature of this tradeoff. They observe that rather than the tradeoff being driven directly by metabolic competition, it appears to occur upstream through regulatory processes including antagonism between hormones. The authors describe situations in which the availability of nutrients or competition shifts the balance between growth and defense, and also strategies of defense regulation that can minimize the negative impact on growth, including priming, transgenerational defense induction, and the fine regulation of R gene expression. Finally, the authors discuss the contributions of other organisms including beneficial insects and the microbiome to low-cost defense strategies. They conclude by emphasizing the need for more field studies to explore growth-defense tradeoffs in “in a range of conditions, and in environments with other species.” Plant Cell 10.1105/tpc.16.00931 Tags: Biotic Interactions, Growth Regulation, Metabolism
It is widely recognized that defense incurs a cost in terms of reduced growth. Karasov et al. explore the nature of this tradeoff. They observe that rather than the tradeoff being driven directly by metabolic competition, it appears to occur upstream through regulatory processes including antagonism between hormones. The authors describe situations in which the availability of nutrients or competition shifts the balance between growth and defense, and also strategies of defense regulation that can minimize the negative impact on growth, including priming, transgenerational defense induction, and the fine regulation of R gene expression. Finally, the authors discuss the contributions of other organisms including beneficial insects and the microbiome to low-cost defense strategies. They conclude by emphasizing the need for more field studies to explore growth-defense tradeoffs in “in a range of conditions, and in environments with other species.” Plant Cell 10.1105/tpc.16.00931 Tags: Biotic Interactions, Growth Regulation, Metabolism
Review: Chloroplast function revealed through analysis of GreenCut2 genes
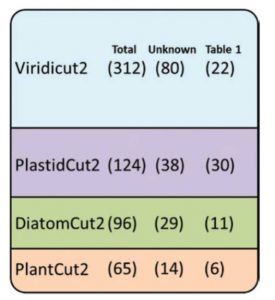 Of the 3000 or so proteins housed in the chloroplast, we know the functions of only a few hundred. One approach to identify function is to first identify plastid proteins found exclusively in photosynthetic organisms. This subset, GreenCut2, is further subdivided by whether the proteins are found in red algae, diatoms, and/or green plants. Fristedt describes further characterization of this set of proteins through literature and database reviews, which led to the sub-categorization of these proteins by their subcellular localization, as well as phylogenetic studies using CLIME (Clustering by inferred models of evolution), which together shed light onto the functions of some of these important but not fully understood proteins. J. Exp. Bot. 10.1093/jxb/erx082 Tags: Bioenergetics, Genomics
Of the 3000 or so proteins housed in the chloroplast, we know the functions of only a few hundred. One approach to identify function is to first identify plastid proteins found exclusively in photosynthetic organisms. This subset, GreenCut2, is further subdivided by whether the proteins are found in red algae, diatoms, and/or green plants. Fristedt describes further characterization of this set of proteins through literature and database reviews, which led to the sub-categorization of these proteins by their subcellular localization, as well as phylogenetic studies using CLIME (Clustering by inferred models of evolution), which together shed light onto the functions of some of these important but not fully understood proteins. J. Exp. Bot. 10.1093/jxb/erx082 Tags: Bioenergetics, Genomics
Review: Many shades of gray – The context-dependent performance of organic agriculture
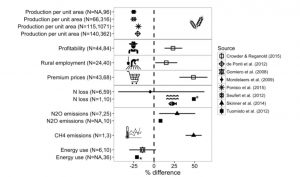 “The benefits of organic agriculture are widely debated. Although some promote it as a solution to our sustainable food security challenges, others condemn it as a backward and romanticized version of agriculture that would lead to hunger and environmental devastation.” Seufert and Ramankutty address these conflicting views through a systematic analysis of organic agriculture’s potential benefits and environmental performance (as affected by lower yields). This is an accessible and information-rich review particularly useful for students. Science Adv. 10.1126/sciadv.1602638 Tags: Applied Plant Biology, Education and Outreach, Environmental Plant Biology
“The benefits of organic agriculture are widely debated. Although some promote it as a solution to our sustainable food security challenges, others condemn it as a backward and romanticized version of agriculture that would lead to hunger and environmental devastation.” Seufert and Ramankutty address these conflicting views through a systematic analysis of organic agriculture’s potential benefits and environmental performance (as affected by lower yields). This is an accessible and information-rich review particularly useful for students. Science Adv. 10.1126/sciadv.1602638 Tags: Applied Plant Biology, Education and Outreach, Environmental Plant Biology
Review: The evo-devo of plant speciation
 Speciation events result from a combination of molecular, environmental and stochastic (random) factors. Several models developed in the last 150 years help to explain how species emerge, but more recently evolutionary developmental biology (evo-devo) approaches give us tools to decipher plant speciation. In this review, Fernández-Mazueco and Glover show us the available evidence, from macro- to micro-evolutionary scales, of why plants are excellent study systems of speciation. They discuss how an integrative study of intra- and inter-specific variation in polyploidy, morphology, phenotypic plasticity, epigenetic inheritance, etc., could shed light on evolution of new species. This is a nice piece of literature for discussing with students! (Summary by Gaby Auge) Nature Ecol. Evol. 10.1038/s41559-017-0110 Tags: Botany, Development, Education and Outreach, Evolution
Speciation events result from a combination of molecular, environmental and stochastic (random) factors. Several models developed in the last 150 years help to explain how species emerge, but more recently evolutionary developmental biology (evo-devo) approaches give us tools to decipher plant speciation. In this review, Fernández-Mazueco and Glover show us the available evidence, from macro- to micro-evolutionary scales, of why plants are excellent study systems of speciation. They discuss how an integrative study of intra- and inter-specific variation in polyploidy, morphology, phenotypic plasticity, epigenetic inheritance, etc., could shed light on evolution of new species. This is a nice piece of literature for discussing with students! (Summary by Gaby Auge) Nature Ecol. Evol. 10.1038/s41559-017-0110 Tags: Botany, Development, Education and Outreach, Evolution
Letter: Picking up the ball at the K/Pg boundary: Ancient polyploidies as a spandrel of asexuality
Roughly 66 million years ago Earth was hit by a huge asteroid, resulting in climate changes that led to mass extinctions, most famously of the non-avian dinosaurs. This catastrophic event, which marks the boundary between the Cretaceous (K) and Paleogene (Pg) periods, also caused widespread mass extinctions in the plant kingdom. Previously, it has been noted that many whole-genome duplications in plants trace to the K/Pg boundary, and a popular selectionist theory is that polyploidization conferred an adaptive advantage under these challenging conditions. Freeling offers an alternative, mutationist hypothesis, that these polyploidizations survived as a byproduct (spandrel) of selection for asexuality: survival of a mass extinction may have depended first on asexuality, and having survived, a polyploid background might then provide an environment of relaxed selection pressure that would support experimentation and absorb massive failure. He concludes by observing the need for a greater understanding of the genetics of asexuality in plants. Plant Cell 10.1105/tpc.16.00836 Tags: Botany, Evolution, Genomics
Commentary: Widespread contamination of Arabidopsis embryo and endosperm transcriptome datasets
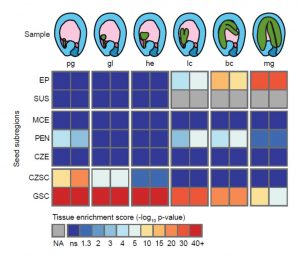 Knowing where a gene is expressed provides valuable information about its function, but that information is compromised if the RNA source is contaminated by other tissues. Schon and Nodine investigated the extent to which Arabidopsis embryo and endosperm transcriptome datasets are affected by tissue contamination. Using seed coat-specific RNAs as markers of maternal tissues and also parent-of-origin small nucleotide polymorphisms (SNPs), the authors found evidence of widespread contamination by maternal tissues, particularly for those datasets representing earlier (smaller) embryos. This finding has implications for interpretations of imprinting (parent-of-origin specific gene expression) in embryos. The authors propose best practices for tissue dissection and also have developed a tool to quantify the extent of contamination of a sample. Plant Cell 10.1105/tpc.16.00845 Tags: Development, Gene Regulation, Molecular Biology
Knowing where a gene is expressed provides valuable information about its function, but that information is compromised if the RNA source is contaminated by other tissues. Schon and Nodine investigated the extent to which Arabidopsis embryo and endosperm transcriptome datasets are affected by tissue contamination. Using seed coat-specific RNAs as markers of maternal tissues and also parent-of-origin small nucleotide polymorphisms (SNPs), the authors found evidence of widespread contamination by maternal tissues, particularly for those datasets representing earlier (smaller) embryos. This finding has implications for interpretations of imprinting (parent-of-origin specific gene expression) in embryos. The authors propose best practices for tissue dissection and also have developed a tool to quantify the extent of contamination of a sample. Plant Cell 10.1105/tpc.16.00845 Tags: Development, Gene Regulation, Molecular Biology
Opinion: Increasing crop yield and resilience with trehalose 6-phosphate ($)
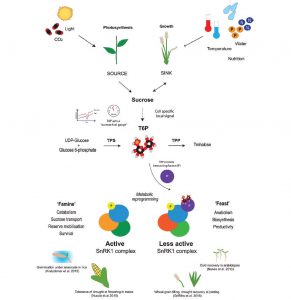 Trehalose 6-phosphate (T6P) is a disaccharide formed from two glucose sugars, and more importantly is a signal of glucose availability and regulator of energy homeostasis. Acting via the protein kinase SnRK1, T6P controls the allocation of carbon, leading the plant down a “feast” (growth) or “famine” (survival) metabolic and growth pathway. Paul et al. review the evidence from three cereal crops that altering T6P levels (through GM, marker-assisted selection and novel chemistry) can promote food security through source-sink optimization, for example by stimulating grain filling even under drought conditions. J. Exp. Bot. 10.1093/jxb/erx083 Tags: Biochemistry, Ecophysiology, Growth Regulation, Metabolism
Trehalose 6-phosphate (T6P) is a disaccharide formed from two glucose sugars, and more importantly is a signal of glucose availability and regulator of energy homeostasis. Acting via the protein kinase SnRK1, T6P controls the allocation of carbon, leading the plant down a “feast” (growth) or “famine” (survival) metabolic and growth pathway. Paul et al. review the evidence from three cereal crops that altering T6P levels (through GM, marker-assisted selection and novel chemistry) can promote food security through source-sink optimization, for example by stimulating grain filling even under drought conditions. J. Exp. Bot. 10.1093/jxb/erx083 Tags: Biochemistry, Ecophysiology, Growth Regulation, Metabolism
Hierarchically aligning 10 legume genomes establishes a family-level genomics platform
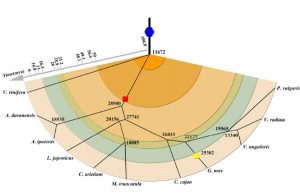 Many legumes are important crops, and to date ten legume genomes have been sequenced, including soybean, common bean, mung bean, and two species of wild peanut. Wang et al. used hierarchical comparative genomics analysis of the ten legume genomes, which enabled them to detected gene colinearity between and within genomes, identify colinearity-supported homologs, orthologs and paralogs, relate duplicated genes to specific ancient polyploidization, and characterize the distribution, expansion, and copy-number variations of economically or agriculturally important gene families and regulatory pathways. Additional data and analyses are available at http://www.legumegrp.org/. Plant Physiol. 10.1104/pp.16.01981 Tags: Computational Biology, Evolution, Genomics
Many legumes are important crops, and to date ten legume genomes have been sequenced, including soybean, common bean, mung bean, and two species of wild peanut. Wang et al. used hierarchical comparative genomics analysis of the ten legume genomes, which enabled them to detected gene colinearity between and within genomes, identify colinearity-supported homologs, orthologs and paralogs, relate duplicated genes to specific ancient polyploidization, and characterize the distribution, expansion, and copy-number variations of economically or agriculturally important gene families and regulatory pathways. Additional data and analyses are available at http://www.legumegrp.org/. Plant Physiol. 10.1104/pp.16.01981 Tags: Computational Biology, Evolution, Genomics
Identification of Arabidopsis genic and non-genic promoters
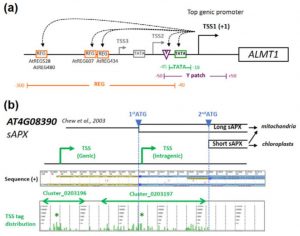 A promoter is a region that “determines the position, direction, frequency, and timing of transcription.” A cell can decode the sequence of the promoter to ensure appropriate transcription, but we still can’t. Tokizawa et al. performed a large-scale survey of promoters by sequencing regions upstream of transcription start sites. They identified promoters corresponding to 80% of Arabidopsis protein-coding genes, as well as many non-coding RNA (miRNA and ncRNA) gene promoters, and intragenic, antisense, and orphan promoters. Onto each of these they mapped core elements (e.g., TATA box) and position-sensitive regulatory elements (REGs). In addition to providing a valuable resource, key findings include that genes average three promoters, and that genes encoding organelle-localized proteins often lack TATA boxes in spite of high expression levels. Plant J. 10.1111/tpj.13511 Tags: Computational Biology, Gene Regulation, Genomics, Molecular Biology
A promoter is a region that “determines the position, direction, frequency, and timing of transcription.” A cell can decode the sequence of the promoter to ensure appropriate transcription, but we still can’t. Tokizawa et al. performed a large-scale survey of promoters by sequencing regions upstream of transcription start sites. They identified promoters corresponding to 80% of Arabidopsis protein-coding genes, as well as many non-coding RNA (miRNA and ncRNA) gene promoters, and intragenic, antisense, and orphan promoters. Onto each of these they mapped core elements (e.g., TATA box) and position-sensitive regulatory elements (REGs). In addition to providing a valuable resource, key findings include that genes average three promoters, and that genes encoding organelle-localized proteins often lack TATA boxes in spite of high expression levels. Plant J. 10.1111/tpj.13511 Tags: Computational Biology, Gene Regulation, Genomics, Molecular Biology
TOPLESS mediates brassinosteroid control of shoot boundaries and root meristem development
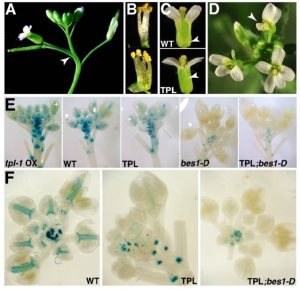 Steroids play a role as essential hormones in plants as well as in animals. In plants, steroids termed brassinosteroids (BR) regulate plant growth and development. BR has been shown to be involved in many processes such as light response, stomata and root development, and flowering in plants. BR-regulated gene expression is controlled by the master regulator bri1-EMS-SUPRESSOR1 (BES1). Espinosa-Ruiz et al. show regulation of organ boundary formation in shoot apical meristem and root quiescent center (QC) quiescence by co-repressors BES1 and TOPLESS. BES1 directs TOPLESS to promoters of their target genes (CUC3 and BRAVO) to suppress their expression. (Summary by Nidhi Sharma) Development 10.1242/dev.143214 Tags: Development, Gene Regulation, Growth Regulation
Steroids play a role as essential hormones in plants as well as in animals. In plants, steroids termed brassinosteroids (BR) regulate plant growth and development. BR has been shown to be involved in many processes such as light response, stomata and root development, and flowering in plants. BR-regulated gene expression is controlled by the master regulator bri1-EMS-SUPRESSOR1 (BES1). Espinosa-Ruiz et al. show regulation of organ boundary formation in shoot apical meristem and root quiescent center (QC) quiescence by co-repressors BES1 and TOPLESS. BES1 directs TOPLESS to promoters of their target genes (CUC3 and BRAVO) to suppress their expression. (Summary by Nidhi Sharma) Development 10.1242/dev.143214 Tags: Development, Gene Regulation, Growth Regulation
Changes in the chloroplastic CO2 concentration explain much of the observed Kok effect: a model ($)
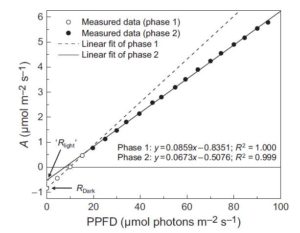 When the uptake of CO2 (A) is plotted against absorbed irradiance (I), at low I there is a noticeable bend that occurs around the light compensation point (I where CO2 release due to mitochondrial respiration is balanced by CO2 uptake by photosynthesis). When the slopes of both parts of the curve are extrapolated back to zero irradiance, it appears as though the rate of mitochondrial respiration is greater in the dark than the light; the inference that there is a light-dependent inhibition of respiration is known as the “Kok effect”. Farquhar and Busch propose a model to explain the Kok effect, in which the apparent curvature of the A vs I plot is attributable to changes in chloroplastic CO2 partial pressure at low I. They suggest that the Kok effect is related to photorespiration, which is consistent with a reduced Kok effect seen in C4 plants, or high CO2 or low O2 conditions. New Phytol. 10.1111/nph.14512 Tags: Bioenergy, Metabolism
When the uptake of CO2 (A) is plotted against absorbed irradiance (I), at low I there is a noticeable bend that occurs around the light compensation point (I where CO2 release due to mitochondrial respiration is balanced by CO2 uptake by photosynthesis). When the slopes of both parts of the curve are extrapolated back to zero irradiance, it appears as though the rate of mitochondrial respiration is greater in the dark than the light; the inference that there is a light-dependent inhibition of respiration is known as the “Kok effect”. Farquhar and Busch propose a model to explain the Kok effect, in which the apparent curvature of the A vs I plot is attributable to changes in chloroplastic CO2 partial pressure at low I. They suggest that the Kok effect is related to photorespiration, which is consistent with a reduced Kok effect seen in C4 plants, or high CO2 or low O2 conditions. New Phytol. 10.1111/nph.14512 Tags: Bioenergy, Metabolism
Shoot-to-root mobile polypeptides involved in systemic regulation of nitrogen acquisition ($)
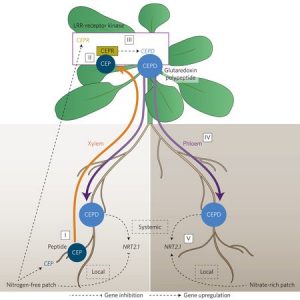 To balance nutrient uptake (usually from heterogeneous sources) with nutrient demand, plants use a root-shoot-root signaling pathway. Previously, a root-to-shoot mobile peptide C-TERMINALLY ENCODED PEPTIDE (CEP) was shown to translocate from N-starved roots to the shoot, where it interacts with a leucine-rich repeat receptor kinase, CEP Receptor 1 (CEPR1). Ohkubo et al. identified two peptides, CEP DOWNSTREAM1 (CEPD1) and CEPD2, that act downstream of CEP-activated CEPR1 and that translocate from shoot to root. CEPD1/2 induce expression of the nitrate transporter NRT2.1 only in roots growing in the presence of nitrate. Therefore, N-starved roots are able to induce nitrate uptake in other roots via this root-shoot-root signalling module: Root-localized CEP sensor detects local N-starvation, ascends to the shoot as a signal, where it induces expression and action of CEPD polypeptides that then translocate to all roots, signaling N demand and resulting in the induction of N-uptake. (Summary by Tyra McCray) Nature Plants 10.1038/nplants.2017.29 Tags: Physiology, Signals and Responses, Transport
To balance nutrient uptake (usually from heterogeneous sources) with nutrient demand, plants use a root-shoot-root signaling pathway. Previously, a root-to-shoot mobile peptide C-TERMINALLY ENCODED PEPTIDE (CEP) was shown to translocate from N-starved roots to the shoot, where it interacts with a leucine-rich repeat receptor kinase, CEP Receptor 1 (CEPR1). Ohkubo et al. identified two peptides, CEP DOWNSTREAM1 (CEPD1) and CEPD2, that act downstream of CEP-activated CEPR1 and that translocate from shoot to root. CEPD1/2 induce expression of the nitrate transporter NRT2.1 only in roots growing in the presence of nitrate. Therefore, N-starved roots are able to induce nitrate uptake in other roots via this root-shoot-root signalling module: Root-localized CEP sensor detects local N-starvation, ascends to the shoot as a signal, where it induces expression and action of CEPD polypeptides that then translocate to all roots, signaling N demand and resulting in the induction of N-uptake. (Summary by Tyra McCray) Nature Plants 10.1038/nplants.2017.29 Tags: Physiology, Signals and Responses, Transport
Living on the edge: conservation genetics of seven thermophilous plant species in a high Arctic archipelago
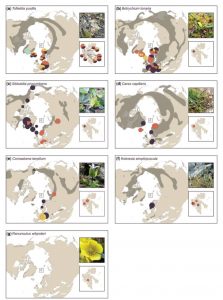 The Arctic provides numerous opportunities to study how climate change and isolation affect plant populations. Birkeland et al. queried the genetic diversity within isolated populations of seven heat-loving (thermophilous) species in the high Arctic (74° – 81° N) Svalbard Archipelago, near the well-known seedbank. These thermophilous plants are thought to be relicts from early Holocene warm periods (9000 – 5000 years ago). The populations studied show low genetic diversity, probably due to founder or bottleneck effects and limited gene flow. This low genetic diversity may limit the ability of these rare plants to benefit from ongoing Arctic warming. AoB Plants 10.1093/aobpla/plx001 Tags: Botany, Evolution, Genetics
The Arctic provides numerous opportunities to study how climate change and isolation affect plant populations. Birkeland et al. queried the genetic diversity within isolated populations of seven heat-loving (thermophilous) species in the high Arctic (74° – 81° N) Svalbard Archipelago, near the well-known seedbank. These thermophilous plants are thought to be relicts from early Holocene warm periods (9000 – 5000 years ago). The populations studied show low genetic diversity, probably due to founder or bottleneck effects and limited gene flow. This low genetic diversity may limit the ability of these rare plants to benefit from ongoing Arctic warming. AoB Plants 10.1093/aobpla/plx001 Tags: Botany, Evolution, Genetics
Canopy near-infrared reflectance and terrestrial photosynthesis
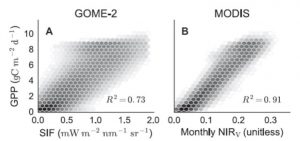 A model is only as good as the data that go into it (garbage in, garbage out), so any effort to improve remote sensing data will contribute to better global models. Badgley et al. describe a new parameter, near-infrared reflectance of vegetation (NIRV), that more accurately quantifies photosynthesis at the global scale (Gross Primary Productivity, GPP) from satellite sensor data. NIRV combines total near-infrared reflectance (NIRT) and the normalized difference vegetation index (NDVI), so therefore “NIRV represents the proportion of pixel reflectance attributable to the vegetation in the pixel.” The authors show that NIRV shows a closer correlation to GPP than solar-induced chlorophyll fluorescence (SIF). Science Adv. 10.1126/sciadv.1602244 Tags: Bioenergetics, Environmental Plant Biology
A model is only as good as the data that go into it (garbage in, garbage out), so any effort to improve remote sensing data will contribute to better global models. Badgley et al. describe a new parameter, near-infrared reflectance of vegetation (NIRV), that more accurately quantifies photosynthesis at the global scale (Gross Primary Productivity, GPP) from satellite sensor data. NIRV combines total near-infrared reflectance (NIRT) and the normalized difference vegetation index (NDVI), so therefore “NIRV represents the proportion of pixel reflectance attributable to the vegetation in the pixel.” The authors show that NIRV shows a closer correlation to GPP than solar-induced chlorophyll fluorescence (SIF). Science Adv. 10.1126/sciadv.1602244 Tags: Bioenergetics, Environmental Plant Biology
Decreasing readability in scientific papers over time
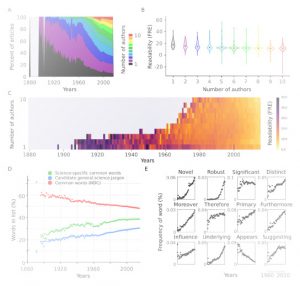 “Reporting science clearly and accurately is a fundamental part of the scientific process, facilitating both the dissemination of knowledge and reproducibility of results.” In this way, Plavén-Sigray et al. introduce us to their preprint in which they analyzed readability in over 700,000 abstracts of scientific articles published in 122 biomedical journals from 1881-2015. Using two different metrics that account for complexity of words and sentences, they found that readability of scientific papers is decreasing steadily over time. This decline is positively correlated with complexity of the rest of the manuscript, showing this trend exceeds abstracts and reflects the whole texts. Moreover, complexity over time is also associated with increasing number of co-authors (although that doesn’t fully explain the trend) and use of in-group science-specific words (jargon). Decreasing readability is a problem when science needs to be explained to non-specialists (digested facts and scientific results can be misinterpreted). This problem even affects specialists, as previously the number of citations has been negatively associated with increasing complexity of language use in papers. So, are we making science less accessible even for ourselves, scientists? (Summary by Gaby Auge) bioRxiv 10.1101/119370 Tags: Education and Outreach
“Reporting science clearly and accurately is a fundamental part of the scientific process, facilitating both the dissemination of knowledge and reproducibility of results.” In this way, Plavén-Sigray et al. introduce us to their preprint in which they analyzed readability in over 700,000 abstracts of scientific articles published in 122 biomedical journals from 1881-2015. Using two different metrics that account for complexity of words and sentences, they found that readability of scientific papers is decreasing steadily over time. This decline is positively correlated with complexity of the rest of the manuscript, showing this trend exceeds abstracts and reflects the whole texts. Moreover, complexity over time is also associated with increasing number of co-authors (although that doesn’t fully explain the trend) and use of in-group science-specific words (jargon). Decreasing readability is a problem when science needs to be explained to non-specialists (digested facts and scientific results can be misinterpreted). This problem even affects specialists, as previously the number of citations has been negatively associated with increasing complexity of language use in papers. So, are we making science less accessible even for ourselves, scientists? (Summary by Gaby Auge) bioRxiv 10.1101/119370 Tags: Education and Outreach
Academic research in the 21st century: Maintaining scientific integrity in a climate of perverse incentives and hypercompetition
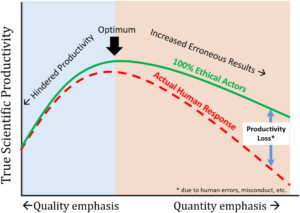 In recent years it has become clear that misconduct in the scientific community is pervasive. In this recent publication by Edwards et al., the authors investigate the potential causes of this problem. Their conclusion is that due to decreased research funding from governments, development of quantitative metrics for research output, and a changing business model for higher education. a climate of hypercompetition is created. As a result, to survive in the modern science community, more and more scientist compromise on their scientific integrity to stay ahead of the competition. (Summary by Harrie van Erp). Env. Eng. Sci. 10.1089/ees.2016.0223 Tags: Education and Outreach, Policy
In recent years it has become clear that misconduct in the scientific community is pervasive. In this recent publication by Edwards et al., the authors investigate the potential causes of this problem. Their conclusion is that due to decreased research funding from governments, development of quantitative metrics for research output, and a changing business model for higher education. a climate of hypercompetition is created. As a result, to survive in the modern science community, more and more scientist compromise on their scientific integrity to stay ahead of the competition. (Summary by Harrie van Erp). Env. Eng. Sci. 10.1089/ees.2016.0223 Tags: Education and Outreach, Policy
