Metabolism is a widely-used term. It is used colloquially, as in, “My metabolism slowed down once I hit 40,” a usage that considers metabolism as a black box and focuses on its energetic inputs and outputs. Or, there’s the single-reaction view of metabolism, as in, “He doesn’t metabolize alcohol very well.” This view underpins the development of pharmaceutical drugs; single molecules that affect single processes.

Metabolism is more than energetics, and more than a collection of reactions. It is both a parts list (metabolite, enzyme, pathway) and the programs that instructs and coordinate the activities of the parts (redox balance, allosteric regulator, transporter). Deconstructing metabolism requires identifying all of the parts and all of the programs.
The first step in understanding metabolism is to construct a pathway diagram, ideally one that shows every metabolite and every enzyme that acts on it, like this one from Roche http://biochemical-pathways.com/#/map/1. Plant scientists owe much to those who study other organisms for their efforts in pathway elucidation, but plants have enormous metabolic capabilities that go beyond E. coli, yeast or Homo sapiens (Moghe and Last, 2015). In spite of much effort, “the functions of at least 60% of predicted Arabidopsis enzymes and transporters remain unclear or unknown” (Niehaus et al., 2015).
Even an elegantly drawn biochemical pathway cannot capture all that is metabolism; we need to know the flux through these pathways and more importantly how the flux changes (Nikoloski et al., 2015). Metabolites have to be considered within the context of the processes they contribute to. In addition to their roles as substrate, product and allosteric regulators, we need to consider their effects on redox balance and charge, their transporters, and the compartmentalization and regulation of enzymes and associated genes that act on them. Genetics, reverse genetics, biochemistry, metabolomics, proteomics and modeling all contribute to our understanding of metabolic dynamics.
Plant breeders have mucked with metabolism since before written history: Seed and fruit size (Doebley et al., 2006), Mendel’s smooth and wrinkled peas (Reid and Ross, 2011), Borlaug’s dwarf wheat (Peng et al., 1999), and black rice (Oikawa et al., 2015) are just a few examples. We’re approaching a very different era in plant breeding, in which we can expect to program the products we obtain from plants (grains, fibers, medicines) by rational changes in the plant’s metabolism by way of gene or genome editing. To realize the potential of predictive biology, we have to know metabolism.
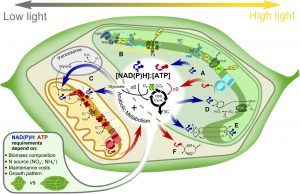
Plant Physiology’s November 2015 Focus Issue on Metabolism, edited by Alisdair Fernie and Eran Pichersky, brings together review and research articles that explore metabolism using bottom up (genomes and proteomes) and top down (traits) approaches. Many of these articles highlight the insights that come from combining methodologies, for example by integrating metabolomics and genomic or transcriptomic data, or by comparing plant and microbial data, or data from plants with different metabolic attributes (see for example Kim and Buell, 2015; Soltis and Kliebenstein, 2015; Toghe et al., 2015).
Metabolism is not fully out of the black box era, but as made evident by this collection we are well on our way to illuminating its workings. As the editors reflect, these articles, “document the extraordinary reach of plant metabolism as well as the application of contemporary approaches being leveraged in order to gain mechanistic insights into both the regulatory processes governing metabolism and the metabolic regulation of a plethora of biological processes,” (Fernie and Pichersky, 2015). This is truly an inspiring collection that doesn’t minimize the challenges of understanding plant metabolism, but at the same time inspires us to explore, seek out and boldly go towards the final frontier of metabolism.
References:
Doebley, J.F., Gaut, B.S. and Smith, B.D. (2006). The Molecular Genetics of Crop Domestication. Cell. 127: 1309-1321.
Fernie, A.R. and Pichersky, E. (2015). Focus Issue on Metabolism: Metabolites, Metabolites Everywhere. Plant Physiology. 169: 1421-1423.
Kim, J. and Buell, C.R. (2015). A Revolution in Plant Metabolism: Genome-Enabled Pathway Discovery. Plant Physiology. 169: 1532-1539.
Moghe, G.D. and Last, R.L. (2015). Something Old, Something New: Conserved Enzymes and the Evolution of Novelty in Plant Specialized Metabolism. Plant Physiology. 169: 1512-1523.
Niehaus, T.D., Thamm, A.M.K., de Crécy-Lagard, V. and Hanson, A.D. (2015). Proteins of Unknown Biochemical Function: A Persistent Problem and a Roadmap to Help Overcome It. Plant Physiology. 169: 1436-1442.
Nikoloski, Z., Perez-Storey, R. and Sweetlove, L.J. (2015). Inference and Prediction of Metabolic Network Fluxes. Plant Physiology. 169: 1443-1455.
Oikawa, T., Maeda, H., Oguchi, T., Yamaguchi, T., Tanabe, N., Ebana, K., Yano, M., Ebitani, T. and Izawa, T. (2015). The Birth of a Black Rice Gene and Its Local Spread by Introgression. Plant Cell. 27: 2401-2414.
Peng, J., Richards, D.E., Hartley, N.M., Murphy, G.P., Devos, K.M., Flintham, J.E., Beales, J., Fish, L.J., Worland, A.J., Pelica, F., Sudhakar, D., Christou, P., Snape, J.W., Gale, M.D. and Harberd, N.P. (1999). `Green revolution/’ genes encode mutant gibberellin response modulators. Nature. 400: 256-261.
Reid, J.B. and Ross, J.J. (2011). Mendel’s Genes: Toward a Full Molecular Characterization. Genetics. 189: 3-10.
Soltis, N.E. and Kliebenstein, D.J. (2015). Natural Variation of Plant Metabolism: Genetic Mechanisms, Interpretive Caveats, and Evolutionary and Mechanistic Insights. Plant Physiology. 169: 1456-1468.
Tohge, T., Scossa, F. and Fernie, A.R. (2015). Integrative Approaches to Enhance Understanding of Plant Metabolic Pathway Structure and Regulation. Plant Physiology. 169: 1499-1511.

1 thought on “Metabolism: The final frontier”