In a study reminiscent of Jurassic Park, scientists have resurrected an extinct enzyme and watched it respond to today’s world. Rubisco is much older than dinosaurs; it is an ancient enzyme, billions of years old, and predates the origin of eukaryotes. Rubisco is found in all photosynthetic organisms, be they bacteria, algae or plant, as well as some chemoautotrophic bacteria that use chemical energy to fix carbon. Therefore, the structure, sequence, and catalytic properties of Rubisco are known for a wide variety of rather distantly related organisms.
The underlying questions of this research are 1) to understand how environmental factors have influenced and constrained Rubisco’s properties, and 2) to what extent those properties can be reverse engineered to make a more efficient enzyme. This is a key question because Rubisco is well known to be a poor enzyme. Not only is it an inefficient catalyst, it also is prone to a detrimental side reaction, as revealed by its name, Ribulose-bisphosphate (RuBP) carboxylase / oxygenase. Its “good” activity is the carboxylation of RuBP, which is the first step in the carbon-fixing Calvin-Benson cycle. Its “bad” reaction is the oxygenation of RuBP, leading to the production of 2-phosphoglycolate which is eventually salvaged but with the expenditure of energy. Thus, the oxygenation activity is a wasteful competitive reaction to the carboxylation activity. The balance between the two activities is affected by the relative amounts of CO2 and O2 at the active site of the enzyme.
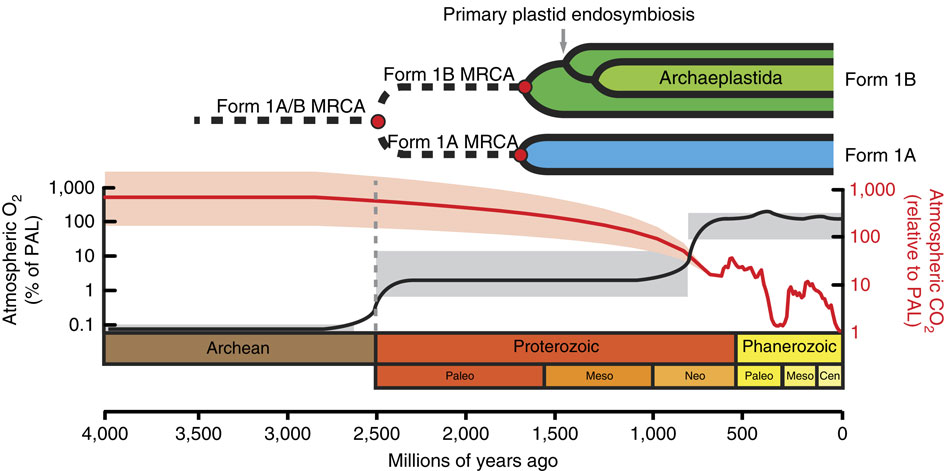
Reconstructing the evolutionary trajectory of Rubisco is particularly interesting because when it evolved, the atmosphere was considerably richer in CO2 and poorer in O2 than present atmospheric conditions, meaning that the oxygenation reaction was not a particular consideration at the time (Figure 1). Over the ensuing billions of years, as atmospheric CO2 levels have dropped (until very recently) and O2 levels have climbed, Rubisco’s oxygenation activity has become more and more significant.
Present day Rubisco is found in many forms. Form IB, the type found in plants and some cyanobacteria, is formed as a hexadecamer of eight each of the large and small subunits. Forms IA, IC and ID have similar L8S8 structures and are found in some cyanobacteria and some proteobacteria (IA), some proteobacteria (IC), and some nongreen algae (ID). Forms II and III lack the small subunit, and are made of various numbers of large-subunit dimers.
There are clear differences in the kinetic properties between prokaryotic and eukaryotic enzymes, in that those of eukaryotes tend to have a higher specificity for CO2 relative to O2 (measured as the ratio of the catalytic efficiency of carboxylation to oxygenation), but a slower reaction rate than those of the prokaryotes, many of which have carbon-concentrating mechanisms that raise the relative CO2 level at the enzyme’s active site. Given the differences in extant Rubisco, and considering the very different environment in which Rubisco evolved, it is reasonable to ask how the ancestral enzyme would compare to today’s enzymes.
By comparing sequences from diverse organisms, Shih et al. were able to project backwards to likely sequences of the large and small subunits for the most recent common ancestor (MRCA) of all Form IA Rubiscos, as well as of all Form IB Rubiscos. The reconstructed sequences were synthesized, the proteins expressed in E. coli, and the holoenzymes assembled and purified for kinetic studies alongside representatives of extant Form1A and 1B enzymes.
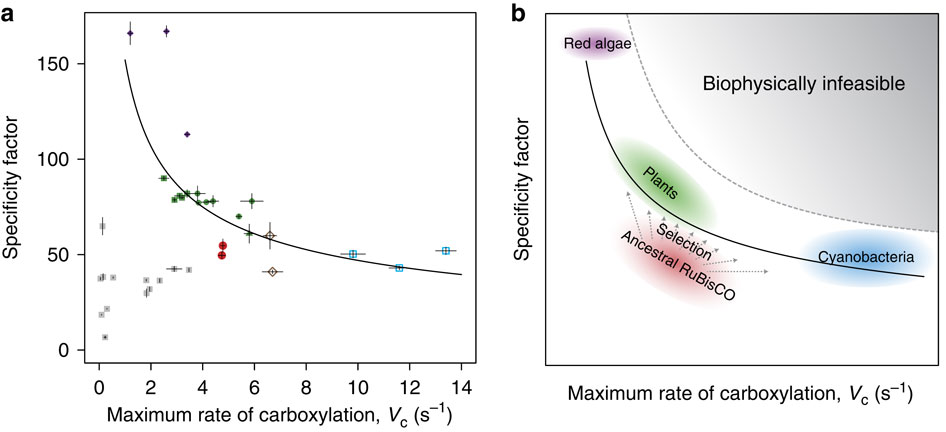
Analysis of the ancestral and extant enzymes indicates that the ancestral enzymes (red circles in Figure 2A) were poorer enzymes than those present today, showing lower rates of catalysis than those of the cyanobacterial enzymes (blue squares in Figure 2A), and lower specificity factors than those of eukaryotic enzymes (green plants = green circles and squares; red algae = purple circles). The ancestral enzymes have apparently evolved in various directions, depending on their local context, as shown in Figure 2B.
Previous efforts to engineer Rubisco towards greater efficiency have been largely unsuccessful, and, interestingly, the reconstructed ancestral enzymes perform better than several of these engineered enzymes (grey squares in Figure 2A). The authors propose that today’s enzymes may be “stalled in a local optimum of the protein fitness landscape,” unable to easily move towards more efficient forms. Therefore, this “ancestral sequence reconstruction provides a unique platform to go back in time, opening the possibilities of forward and reverse engineering on an enzyme that has not yet been subjected to the selective pressures of history.”
Steven Spielberg may not be able to make a blockbuster movie from this work, but giving life another chance to find a way is still pretty exciting stuff.
Shih P.M., Occhialini A., Cameron J.C., Andralojc P.J., Parry M.A.J.and Kerfeld C.A. (2016). Biochemical characterization of predicted Precambrian RuBisCO. Nat Commun 7: 10382.
