Guest post from Sonja Dunbar, PhD student at the University of Cambridge
Plants need nutrients and they have a lot of different ways to acquire them from the soil, as the 2nd year undergraduate students I teach at the University of Cambridge recently discovered in lectures. One thing our lecture courses try to emphasise at this stage of their degree is the importance of experimental evidence supporting the facts we teach. However, getting to grips with the different experiments isn’t always easy when you can’t try them out for yourself.
To try and make experiments more accessible, I developed the following activity for use in small group teaching (2-4 students, although it might be adaptable for larger groups). The aim of this exercise was to give the students a real world problem and get them to design a series of experiments to investigate. I used the figures from Chao et al., 2014 to provide them with the data from these experiments, allowing them to interpret the data and decide on the next course of action. This way we were able to build up a thorough investigation and show the students that using their theory they were capable of designing a publishable course of research.
The students were introduced to the activity with the following setup:
“Arsenic is a problem in many parts of the world. Where groundwater contains high levels of arsenic it is taken up by plants and builds up in plant matter, leading to increased levels of arsenic in the food chain. This occurs because arsenate, the common form of arsenic in the environment, is chemically similar to phosphate and is accidentally taken into the plant by phosphate transporters.
We know that plants are able to detoxify arsenate by converting it to arsenite which can then be extruded back into the environment. Your task is to identify a gene involved in arsenate detoxification and show that the product it encodes is critical to arsenate elimination. This information would then allow us to look into improving arsenic tolerance in key crops, particularly rice.
To investigate the arsenic tolerance gene you have available to you a library of 349 ecotypes of Arabidopsis thaliana and standard laboratory equipment for plant growth and DNA/protein analysis. What do you do?”
Most groups might need a bit of prompting to get to grips with this activity at the beginning as it is quite different to usual supervision activities!
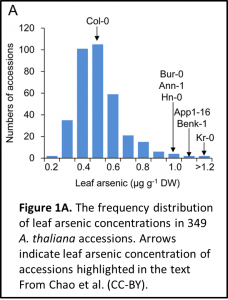
Once they have the idea that they should challenge the ecotypes with arsenic, I show them Figure 1A for interpretation. This figure shows the amount of leaf arsenic accumulated in different ecotypes (natural variants). Having identified Kr-0 as a good ecotype for this investigation due to its high levels of arsenic, I summarise the findings of the genome wide association mapping and validation of the locus (Figure 2). 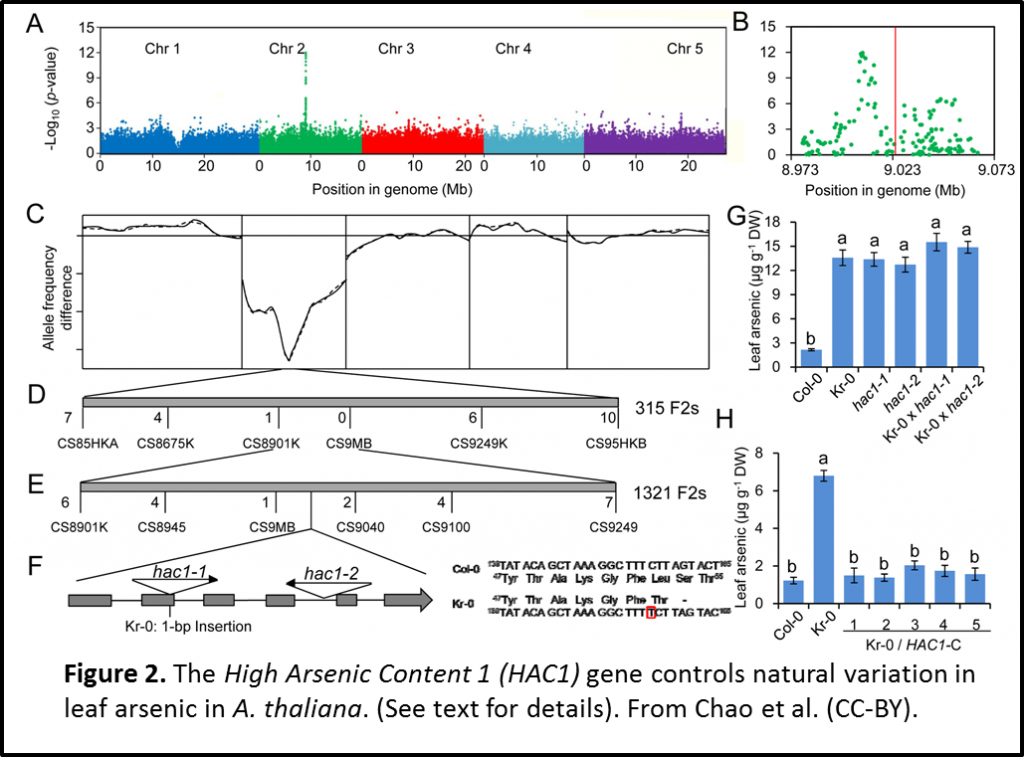
I then challenged them to investigate this gene with methods they are familiar with from the lectures including DNA sequencing, genetic complementation and expression studies and reciprocal grafting . At this point it is likely you will jump around the different figures to provide results for them – I found figures 3, 4, 6 and 7 particularly useful and easy for them to interpret.
If the students are struggling with ideas, you could try showing them a figure and getting them to work backwards from there. I did this with figure 5 to introduce the idea of looking for similar sequence motifs across other species for hints as to the function of this gene.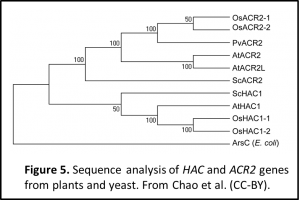
Going through the evidence to demonstrate that the gene is an arsenate reductase, expressed in the roots and induced by arsenate took approximately 20 minutes. I concluded the activity there due to time constraints, but felt that we could have continued to further investigate its role using figures 8-10 as the students were still enthusiastic.
At the end of each supervision I ask the students to write down a key thing they learnt today. Despite there being several other activities in the session, the arsenic activity featured highly as you can see in a selection of their comments. Chatting to them also showed that they were particularly excited to see how you could design a set of experiments to get from question to answer and they were chuffed to be able to have come up with a publishable experiment plan! So, if you would like to get your students to see the relevance of the experimental evidence they see in lectures, I’d recommend giving this a go!
Reference
Chao, D.-Y., Chen, Y., Chen, J., Shi, S., Chen, Z., Wang, C., Danku, J.M., Zhao, F.-J. and Salt, D.E. (2014). Genome-wide Association Mapping Identifies a New Arsenate Reductase Enzyme Critical for Limiting Arsenic Accumulation in Plants. PLoS Biol. 12: e1002009.
Sonja Dunbar is a PhD candidate in the laboratory of Dr Laura Itzhaki at the University of Cambridge. She studies protein-protein interactions using biophysical methods. Her love of plants stems from her degree and she can be found talking about them on Twitter (@PlantSciSonja), on SciCam (https://www.youtube.com/user/SciCamVideos) and in her teaching and outreach. When not in the lab, Sonja enjoys boardgames and gardening (with a lot of tips from her parents!).
Do you lead a guided inquiry of a research paper with undergraduates and would you be willing to share your strategy? Drop me a line if you are interested in helping others help students engage with the literature! mwilliams@aspb.org.