Scientists have long dreamed of the ability to make targeted genomic changes: precise and specific alterations in an organism’s DNA to affect its phenotype. Recently, this dream has become a reality through the discovery and engineering of nucleases that can be targeted to precise genomic locations.
Double-strand breaks produced by targeted nucleases can induce mutations or provide the opportunity for DNA insertions or deletions. The first types of targeted nucleases included zinc-finger nucleases (ZFNs) and transcription-activator-like effector nucleases (TALENs), which are targeted via programmable DNA-binding domains of proteins (for reviews see Carroll, 2014; Kim and Kim, 2014; Puchta and Fauser, 2014; Voytas, 2013; Weeks et al., 2015).
More recently, the CRISPR/Cas9 method has been developed, which targets the Cas9 nuclease to a DNA locus through an RNA guide. This RNA-based targeting makes CRISPR/Cas9 even more powerful and versatile than systems that use protein-based targeting.
A CRISPR primer
Like restriction endonucleases, CRISPR technology is derived from a prokaryotic defense strategy called the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated (Cas) system. In defense, a short DNA sequence from an invading DNA element such as a virus or plasmid (indicated in red in Figure 1) is incorporated into the CRISPR locus. Transcription of this locus produces CRISPR RNAs (crRNAs) which associate with Cas proteins to target and cleave the invading DNA.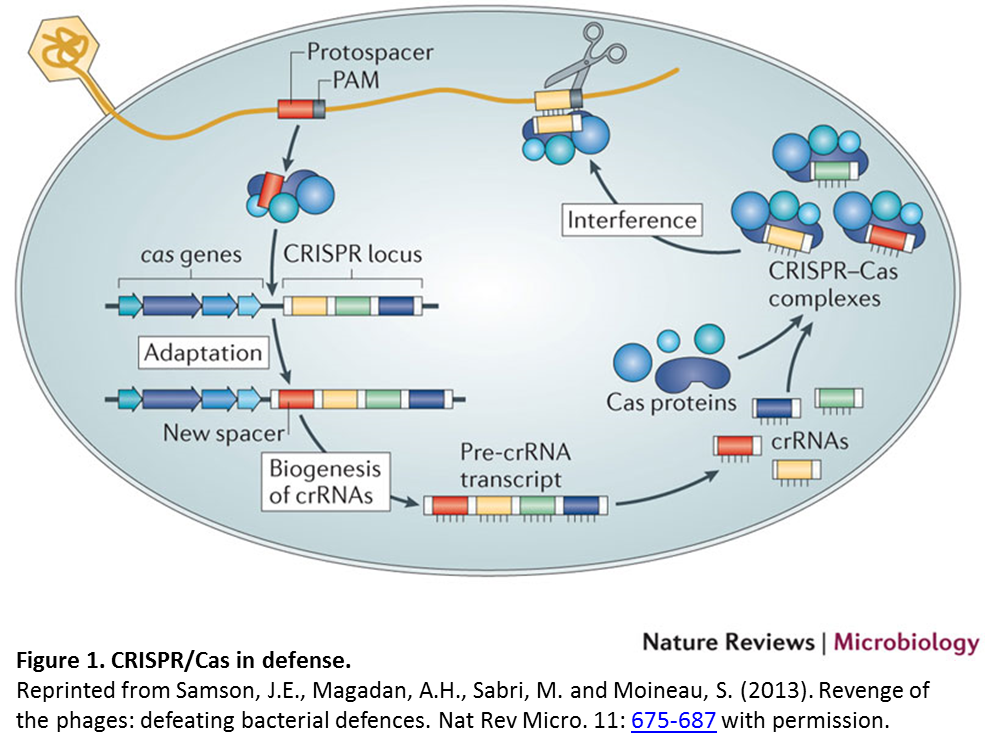
The system has been modified to make it useful for targeted genome editing, via expression of a single-guide RNA (sgRNA). By judicious sgRNA design 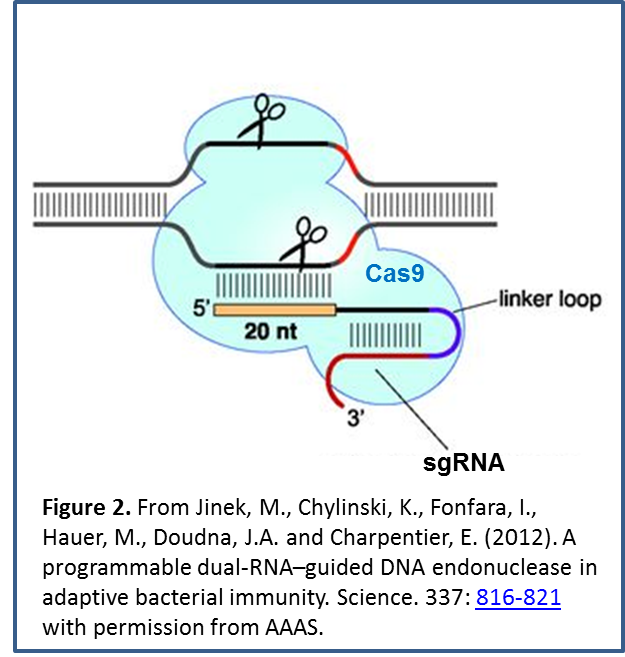 (Figure 2), nucleases (or other proteins) can be directed to specific genomic regions. Double-strand DNA breaks produced by nucleases can be improperly repaired to knock out gene function, or the site can incorporate new DNA sequences provided by a template to replace or add a gene.
(Figure 2), nucleases (or other proteins) can be directed to specific genomic regions. Double-strand DNA breaks produced by nucleases can be improperly repaired to knock out gene function, or the site can incorporate new DNA sequences provided by a template to replace or add a gene.
Application of CRISPR/Cas9 for genome editing
The potential of CRISPR/Cas9 technology in plants has been well demonstrated (see Baltes and Voytas, 2015; Belhaj et al., 2013; Belhaj et al., 2015; Bortesi and Fischer, 2015; Ma et al., 2015; Puchta and Fauser, 2014; Xing et al., 2014; and references therein). Furthermore, CRISPR-based RNA-targeting can be harnessed for a wider range of genome edits (Sternberg and Doudna, 2015). For example, eliminating the nuclease activity from Cas9 (resulting in dCas9) enables it to serve as a DNA-binding domain that can anchor other proteins to specific sites, enabling CRISPR/Cas9 to be a site-specific system for targeting diverse proteins to specific genes. In a recent paper in Plant Physiology, Lowder et al., (2015), introduce a toolbox for CRISPR/Cas9 methods in plants, including a variety of applications as described in Figure 3.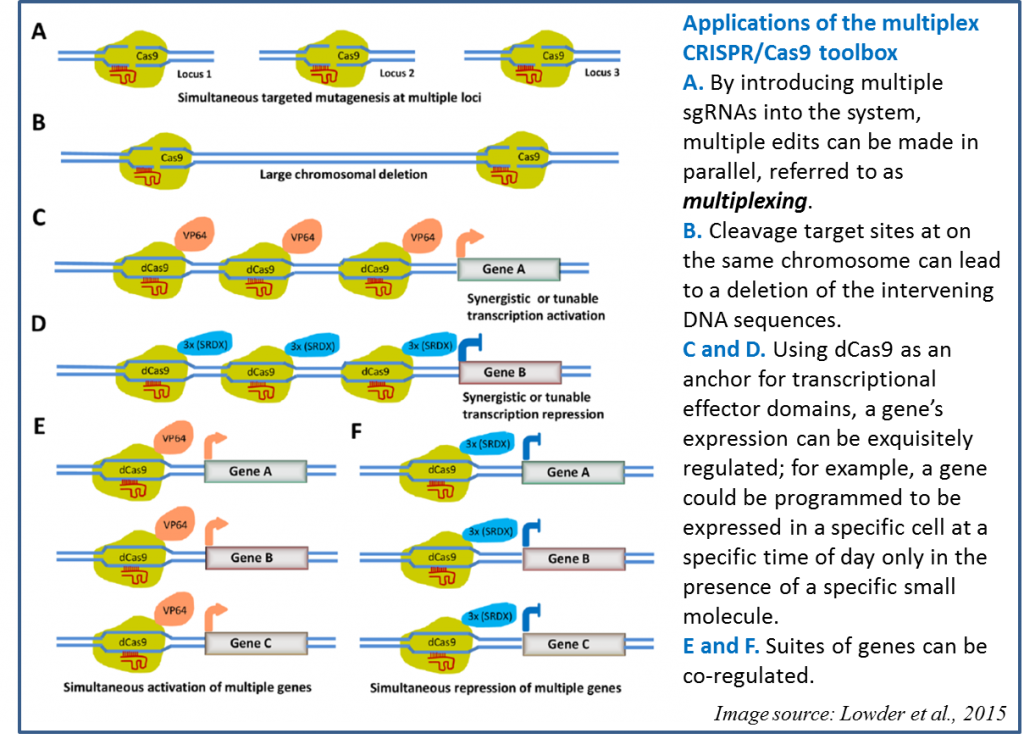
The power of CRISPR/Cas9 has renewed discussions on the ethical implications of human embryo engineering and the release of engineered wild animals (e.g., is it a “biocontrol silver bullet or global conservation threat?”). For plant scientists, the technology may provide a work-around for the public relations nightmare that plagues transgenic crop improvement efforts, thereby increasing the speed of introduction of new traits (see Araki and Ishii, 2015; Voytas and Gao, 2014). Some future applications are obvious, for example knocking out the function of an undesirable gene. Others applications depend upon a more sophisticated understanding of gene functions and regulatory networks; however, the development of CRISPR/Cas9 as method should greatly increase the speed at which we acquire this knowledge by facilitating precise genome engineering. One thing seems sure though, that CRISPR/Cas9 has the potential to revolutionize molecular biology as thoroughly as did restriction enzymes in the 1970s and PCR in the 1980s/1990s.
References
Araki, M. and Ishii, T. (2015). Towards social acceptance of plant breeding by genome editing. Trends Plant Sci. 20: 145-149.
Baltes, N.J. and Voytas, D.F. (2015). Enabling plant synthetic biology through genome engineering. Trends Biotechnol. 33: 120-131.
Belhaj, K., Chaparro-Garcia, A., Kamoun, S., Patron, N.J. and Nekrasov, V. (2013). Plant genome editing made easy: targeted mutagenesis in model and crop plants using the CRISPR/Cas system. Plant Methods 9: 39.
Belhaj, K., Chaparro-Garcia, A., Kamoun, S., Patron, N.J. and Nekrasov, V. (2015). Editing plant genomes with CRISPR/Cas9. Curr. Opin. Biotechnol. 32: 76-84.
Bortesi, L. and Fischer, R. (2015). The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol. Adv. 33: 41-52.
Carroll, D. (2014). Genome engineering with targetable nucleases. Annu. Rev. Biochem. 83: 409-439.
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J.A. and Charpentier, E. (2012). A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science. 337: 816-821
Kim, H. and Kim, J.-S. (2014). A guide to genome engineering with programmable nucleases. Nat. Rev. Genet. 15: 321-334.
Lowder, L.G., Zhang, Y., Baltes, N.J., Paul, J.W., Tang, X., Zheng, X., Voytas, D.F., Hsieh, T.-F., Zhang, D. and Qi, Y. (2015). A CRISPR/Cas9 toolbox for multiplexed plant genome editing and transcriptional regulation. Plant Physiol. In press. http://www.plantphysiol.org/content/early/2015/08/25/pp.15.00636.abstract.
Ma, X., Zhang, Q., Zhu, Q., Liu, W., Chen, Y., Qiu, R., Wang, B., Yang, Z., Li, H., Lin, Y., Xie, Y., Shen, R., Chen, S., Wang, Z., Chen, Y., Guo, J., Chen, L., Zhao, X., Dong, Z. and Liu, Y.-G. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol.Plant. 8: 1274-1284.
Puchta, H. and Fauser, F. (2014). Synthetic nucleases for genome engineering in plants: prospects for a bright future. Plant J. 78: 727-741.
Samson, J.E., Magadan, A.H., Sabri, M. and Moineau, S. (2013). Revenge of the phages: defeating bacterial defences. Nat Rev Micro. 11: 675-687.
Sternberg, Samuel H. and Doudna, Jennifer A. (2015). Expanding the biologist’s toolkit with CRISPR-Cas9. Mol. Cell. 58: 568-574.
Voytas, D.F. and Gao, C. (2014). Precision genome engineering and agriculture: opportunities and regulatory challenges. PLoS Biol. 12: e1001877.
Voytas, D.F. (2013). Plant genome engineering with sequence-specific nucleases. Annu. Rev. Plant Biol. 64: 327-350.
Weeks, D.P., Spalding, M.H. and Yang, B. (2015). Use of designer nucleases for targeted gene and genome editing in plants. Plant Biotechnol. J. In press. http://onlinelibrary.wiley.com/doi/10.1111/pbi.12448/abstract
Xing, H.-L., Dong, L., Wang, Z.-P., Zhang, H.-Y., Han, C.-Y., Liu, B., Wang, X.-C. and Chen, Q.-J. (2014). A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biology. 14: 327.
Update Dec 2015:
Here’s another important reference for this list
Lawrenson, T., Shorinola, O., Stacey, N., Li, C., Ostergaard, L., Patron, N., Uauy, C. and Harwood, W. (2015). Induction of targeted, heritable mutations in barley and Brassica oleracea using RNA-guided Cas9 nuclease. Genome Biology. 16: 258.

1 thought on “CRISPR here, CRISPR there, CRISPR CRISPR everywhere”